

Population genomics of a baboon hybrid zone in Zambia
My dissertation research is centered on understanding the evolutionary dynamics of a common phenomenon in nature: hybridization.
Hybridization is the interbreeding between distinct biological populations. Oftentimes, the term is used to refer to interbreeding between species (i.e., interspecies hybridization) but hybridization can also take place at any level above or below that of the species.
The concept of species is fundamental to biology. Although biologists disagree about the exact definition under various species concepts, they largely agree that the biological world is naturally organized into species units that are separated from other such units by distinct characteristics and ecological roles. They also agree that this organization stems from populations having divergent evolutionary histories due to, at least for sexually reproducing species, an absence of interbreeding between populations.
This emphasis on the ability to interbreed makes hybridization extremely important for understanding speciation in animals. Even if populations already qualify as species under one or more species concepts, the presence or possibility of hybridization (the latter often seen in captive conditions, such as in the “zeedonk” below) signifies that the populations are still undergoing the same process of differentiation that produces species.

Hybridization has another important benefit for biological research. Because species share many features of their appearance, ecology, behavior, genetics, and so on, isolating the causes or mechanisms underlying these features (phenotypes) is challenging because these features are usually present together. Hybridization provides a workaround for this problem by breaking apart these associations beginning from the genetic level. In the laboratory, crosses performed for this purpose are commonly used to deduce or quantify the contribution of genes to phenotypes in model organisms. For non-model organisms that are impractical or unethical to cross in laboratory settings, natural hybridization provides a special opportunity to study the causes of biological variation.
For this project, I am interested in cases of natural hybridization between populations in the process of speciation, particularly in baboons.
Baboons are widespread adaptable primates distributed across sub-Saharan Africa and parts of the Arabian Peninsula. Across this distribution, they inhabit a diverse range of habitats including savannas, deserts, woodlands, rainforests, mountains, and mangroves.
Distribution of baboons (genus Papio)
| ? | ? | ? |
| ? | ? | ? |
Mouse over map to identify countries
As with many similarly widespread animals, baboons have a parapatric distribution, which means that different forms inhabit adjacent areas with narrow zones of overlap (contact zones). These distributions tend to correspond with distinct habitat types and the lines of overlap tend to be located at transition areas between habitats. Contact zones offer a strong potential for hybridization. In fact, baboons appear to hybridize readily when given the opportunity, as evidenced by a number of known cases of ongoing hybridization and by signatures of past hybridization in the genomes of all baboons.
Known cases of natural hybridization in baboons
| Locality | Species | References | ||
|---|---|---|---|---|
| Awash River, Ethiopia | hamadryas baboons olive baboons |
Jolly & Brett 1973 Nagel 1973 Sugawara 1979 Phillips-Conroy et al. 1986 |
||
| Amboseli Basin & Simba-Kiboko, Kenya | olive baboons yellow baboons |
Maples & McKern 1967 Samuels & Altmann 1986 Tung et al. 2008 Charpentier et al. 2012 |
||
| Luangwa Valley, Zambia | chacma baboons Kinda baboons yellow baboons |
Burrell 2009 | ||
| Kafue River Valley, Zambia | chacma baboons Kinda baboons |
Jolly et al. 2011 | ||
| Bole Valley, Ethiopia | geladas (Theropithecus) olive baboons |
Dunbar & Dunbar 1974 | ||
| Tanzania (historical) | kipunji (Rungwecebus) multiple baboons |
Burrell et al. 2009 Zinner et al. 2009 |
||
| Other historical hybridization | multiple baboons |
Wildman et al. 2004 Zinner et al. 2009 Keller et al. 2010 |
From a genetic perspective, hybrid zones are curious areas where alleles (variants of genes) that were previously absent in one population can cross the hybrid zone and subsequently move farther and farther into that population’s range through successive crossings of hybrid individuals with parental (“pure”) populations. This process is known as introgression.
Gene flow across hybrid zones can vary from none (for instance, if hybrid individuals are infertile) to extensive depending on many factors including ecology, demography, climate, geography, and behavior. When gene flow is present, different genomic regions move across the hybrid zone at different rates, with the quickest being those providing an adaptive advantage in the recipient population. By contrast, alleles not providing an adaptive advantage will be less likely to transfer across populations. For this reason, alleles that are favored in local environments but not others (or, alleles under local adaptation) tend to move slowly because of their loss of adaptive advantages outside of optimal local conditions.
Most but not all markers are inherited biparentally (one allele from each parent). For other markers (the sex-linked markers), alleles can show different patterns of introgression due to factors such as sex-biased dispersal behavior, sex-biased mate recognition between species, or the variable viability of male versus female hybrids (the sex with non-matching chromosomes tends to be less viable).
In this project, I use populations genomics to examine hybridization in Zambian baboons, concentrating especially on the social, demographic, and ecological factors influencing patterns of introgression across the hybrid zone.
Hybridization at my study site is between Kinda baboons (Papio kindae) and gray-footed chacma baboons (Papio ursinus griseipes), also known as “grayfoot” baboons.
Comparison of Kinda and grayfoot baboon faces.

Kinda baboons have only recently become recognized as a major form (or species) of baboon in need of study. Many aspects of their anatomy, physiology, ecology, and behavior are just beginning to emerge. These new pieces of evidence together indicate that Kinda baboons have developed a very distinct suite of characteristics, some of which depart significantly from the typical baboon pattern.
Kinda baboons, for instance, are the smallest species of baboons, both in overall size and in the relative size difference between adult males and females (also known as body size sexual dimorphism). They have a slender, gracile build and have a small, rounded cranium with a short muzzle. This set of features is unusual in adult males and is not unlike features found in juveniles and subadults of other baboon species, suggesting that male Kinda baboons may have evolved physiological modifications stunting development.
Comparison of average adult body weight of males and females of six baboon species.
Ongoing behavioral research on Kinda baboons is also indicating that Kinda baboons are peculiar among baboons in some aspects of their behavior. Unusually, adult Kinda males frequently initiate and maintain extended grooming interactions with females even while the females are not in estrus—female baboons, like many Old World primates, have sexual skin that swells cyclically and conspicuously around the period of maximum sexual receptivity. Grooming in primates and other mammals is used as a powerful form of social currency, but its use towards non-estrous females by adult males is unusual and suggests that there are very different social strategies at work in Kinda baboons.

In many Old-World primates, females often utter distinctive low-pitched rhythmic vocalizations during or immediately following copulations with males. These vocalizations have been hypothesized to benefit females by inciting male competition or by encouraging infant protection by male partners. Unlike all other baboons, in Kinda baboons these vocalizations appear to be either absent or especially rare and inconspicuous. This peculiarity again hints that male and female relationships are uniquely different in Kinda baboons.
Copulation calls in baboons
Grayfoot baboons are much more well-known. As with other “savanna baboon” species, grayfoot baboons live in social groups with multiple adult males and females. Females in the group are characterized by strong female bonds that closely mirror kin relationships. Females are on average more closely related than males because of the tendency for females to remain in their birth groups throughout their lives, a pattern known as female philopatry. Young adult males, in contrast, will emigrate (or disperse) from their birth groups upon reaching maturity in search of reproductive opportunities in other groups. In their new groups, males compete with other males for dominant social positions leading to greater access to females. In grayfoot baboons, a few males sire most of the offspring in a group, resulting in high reproductive skew.

Hybridization between Kinda and grayfoot baboons is particularly unusual given the size difference between the two species. The partners in a Kinda male and grayfoot female pairing have similar weights while for a grayfoot male and Kinda female pairing, the male weighs nearly three times as much as the female!
Weight differentials in Kinda/grayfoot baboon hybridization.
Given this dramatic size difference, one might expect that grayfoot males should dominate interspecies reproduction due to their ability to physically outcompete Kinda males. A recent genetic survey, however, found that in fact the opposite is true. Based on Y-chromosome and mitochondrial markers and taking advantage of the fact that the Y-chromosome can only be passed from fathers to sons and the mitochondrial genome from mothers to offspring, the study found (1) that Kinda Y markers were present in groups that were phenotypically grayfoot or mixed, and (2) that all genetically hybrid males (males with Y and mitochondrial markers from different species) carried Kinda Y-chromosomes. Possible explanations for this unexpected finding are that Kinda males possess some social or behavioral advantage over grayfoot males in securing matings with females, or that there may be reproductive incompatibilities specific to grayfoot male and Kinda female pairings.
The goal of this dissertation project is to explore the factors and forces that influence the spatial distribution of genetic variation across hybrid zones. Which genes are transferred across species boundaries? How far do they go? Why do they end up where they are? What does this tell us about speciation and evolution?
In order to address these questions, I am conducting a genetic survey at Kafue National Park, where the distributions of Kinda and grayfoot baboons join up, forming an extended contact zone over which hybridization occurs.
Kafue National Park, Zambia.
This survey will expand on previous work both in terms of map coverage and genome coverage. In fact, the approach used in this study is a genomic approach, which means that instead of evaluating the impacts of hybridization on a few genetic markers, this study evaluates the impacts of hybridization on the genome as a whole.
Study area: Kafue National Park.
As a biological anthropologist, my ultimate goal is to use baboons as a model for human variation and evolution. But what does baboon hybridization have to do with humans? Why not study humans directly?
Indeed, the field of human genomics is extremely active and continues to generate important findings, many of which relate to human variation and evolution. Ultimately, however, human genomics must sample from either living human populations or an extremely limited supply of preserved extinct human specimens for which DNA is intact. This is problematic as, despite their global distribution, living human populations are highly related and have little genetic variation. While humans do show geographical biological variation, some of which is based in genetics, discrete boundaries in variation between populations do not exist (defining races or other groupings biologically is impossible for this reason). Humans also exhibit considerable social and behavioral variation across their geography, but the bulk of this variation is cultural and not biological. Contrary to popular belief in many parts of the world, the social groups that we recognize are socially transmitted historical constructions and not a reflection of genetic variation.
These features make humans poor models for understanding processes of speciation and hybridization. This is especially interesting given that, just 30-200 thousand years, ancestors of modern humans were living side-by-side with more than one population (or species) of extinct humans. Recent genetic research making use of DNA sequenced from ancient specimens has revealed that human ancestors in fact hybridized with at least two extinct populations of “archaic” humans—Neanderthals and Denisovans—resulting in traces of archaic DNA in modern human genomes.
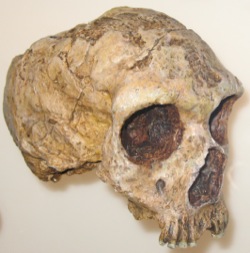
Because they share many features of their evolutionary history, biology, and distribution, baboons are ideal natural models for human hybridization and evolution. Like humans, baboons are widespread, highly social and adaptable, living in a host of different environments. Unlike modern humans, however, baboons do form discrete parapatric species or populations that hybridize at their zones of overlap. They also exhibit population-specific features of their behavior and social organization, some of which are genetically based. Baboons therefore offer exciting opportunities to scientifically evaluate the forces shaping genetic variation at hybrid boundaries while providing powerful analogies for understanding similar processes in human evolution.
One of the first decisions involved in any population genetics project relates to what kind of DNA marker to study, and which. Not all markers need to be sequenced directly down to the level of each DNA base (nucleotide). Microsatellites (DNA regions consisting of multiple repeats of the same short sequence of DNA; the number of repeats varies according to individual), for example, are popular for population genetics because their variation can be easily determined by testing for differently sized regions in different individuals. Similarly, restriction enzymes were popular in the past because they easily and inexpensively generated DNA fragments that differed in size among individuals (see below).
Restriction fragment length polymorphisms (RFLPs).
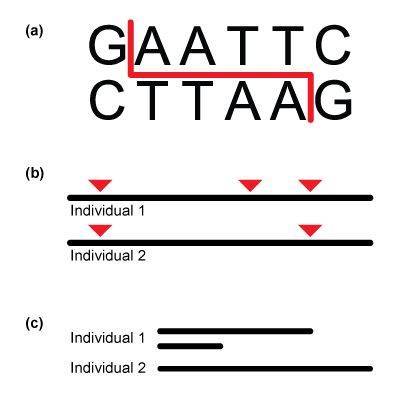
Unlike in microsatellites and RFLPs, the source of genetic variation analyzed for this project will come from variation at the DNA base level, in the form of single nucleotide polymorphisms, or SNPs (commonly pronounced “snips”). SNPs arise due to mutations causing single-base changes (or substitutions) in DNA.
One challenge for analyzing SNPs lies in choosing which SNPs to sequence. One approach would be to sequence whole genomes for multiple individuals within a population and finding the SNPs. This process, however, is time consuming, expensive, and impractical for working with many individuals. Another approach would be to use available genome data to identify a large number of known SNPs, then to use the surrounding sequences to engineer probes that target those regions. This approach, however, requires that the genomes of multiple individuals of a species are already available, a luxury that is almost never present. In fact, the very point of sequencing most organisms is that the data we need do not yet exist!
A recent method overcomes these challenges by targeting many regions across the genome without the need for complete genome sequencing or the availability of existing reference genomes. This method is known as restriction-site-associated DNA sequencing, or RADseq. As its name suggests, restriction enzymes are again involved. In RADseq, only genomic regions surrounding restriction enzyme cut sites are sequenced and searched for SNPs using next-generation sequencing technologies. Restriction enzyme cut sites are scattered throughout the genome and their locations are effectively random, and so this method is able to avoid many issues of statistic bias. Because it provides genomic scale data without the need for sequencing the entire genome, RADseq is part of a family of techniques known as reduced representation techniques. In humans and other primates, RADseq has been used to target tens of thousands of DNA sites, several orders of magnitudes greater than the number of sites that can be sequenced practically using microsatellites.
Because of its many advantages, RADseq will be used in this project to generate genome-wide SNP markers for baboon population genetics. These markers will then be used to infer the demographic history of Kafue National Park baboons and to assess the impacts of behavioral, demographic, and ecological factors on genetic variation across the hybrid zone.
This material is based upon work supported by the National Science Foundation under Grant Number BCS 1341018. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation.